Framework for predicting alloreactivity in hematopoietic cell transplants
Technology tamfitronics
- Research Briefing
- Published:
Nature Biotechnology (2024)Cite this article
Subjects
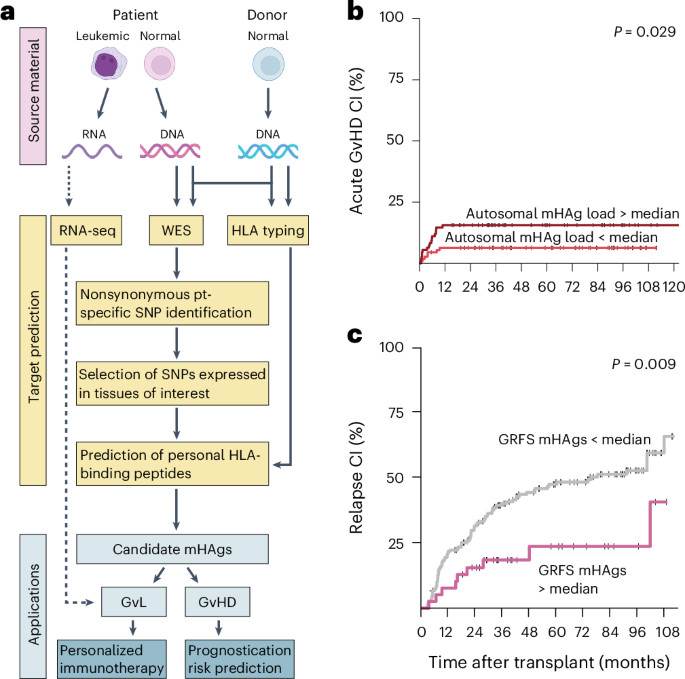
The therapeutic principle of allogeneic hematopoietic cell transplantation, one the most common forms of cancer immunotherapy, is alloreactivity, yet its molecular determinants remain largely unknown. An analytical framework now enables personalized assessment of alloreactivity from whole-exome sequencing of donor–recipient pairs, to help with prognostication of disease relapse and immune-mediated complications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
214,86 € per year
only 17,91 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

References
-
Blazar, B. R., Hill, G. R. & Murphy, W. J. Dissecting the biology of allogeneic HSCT to enhance the GvT effect whilst minimizing GvHD. Nat. Rev. Clin. Oncol. 17475–492 (2020). A review of our current understanding of the GvL effect.
Article PubMed PubMed Central Google Scholar
-
Shlomchik, W. D. Graft-versus-host disease. Nat. Rev. Immunol. 7340–352 (2007). A review that provides mechanistic insights into GvHD immunobiology.
Article CAS PubMed Google Scholar
-
Biernacki, M. A., Sheth, V. S. & Bleakley, M. T cell optimization for graft-versus-leukemia responses. JCI Insight 5e134939 (2020). A review that describes the alloantigenic and antigenic landscapes after transplant as well as strategies to augment the GvL effect.
Article PubMed PubMed Central Google Scholar
-
Griffioen, M., van Bergen, C. A. & Falkenburg, J. H. Autosomal minor histocompatibility antigens: how genetic variants create diversity in immune targets. Front. Immunol. 7100 (2016). A review that provides a comprehensive compendium of known mHAgs.
Article PubMed PubMed Central Google Scholar
-
Mutis, T., Xagara, A. & Spaapen, R. M. The connection between minor H antigens and neoantigens and the missing link in their prediction. Front. Immunol. 111162 (2020). A review that establishes a fascinating parallelism between neoantigens and mHAgs.
Article CAS PubMed PubMed Central Google Scholar
Download references
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Cieri, N. et al. Systematic identification of minor histocompatibility antigens predicts outcomes of allogeneic hematopoietic cell transplantation. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02348-3 (2024).
Rights and permissions
About this article
Cite this article
Framework for predicting alloreactivity in hematopoietic cell transplants. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02354-5
Download citation
-
Published:
-
DOI: https://doi.org/10.1038/s41587-024-02354-5
Technology tamfitronics Associated content
Discover more from Tamfis Nigeria Lmited
Subscribe to get the latest posts sent to your email.



 Hot Deals
Hot Deals Shopfinish
Shopfinish Shop
Shop Appliances
Appliances Babies & Kids
Babies & Kids Best Selling
Best Selling Books
Books Consumer Electronics
Consumer Electronics Furniture
Furniture Home & Kitchen
Home & Kitchen Jewelry
Jewelry Luxury & Beauty
Luxury & Beauty Shoes
Shoes Training & Certifications
Training & Certifications Wears & Clothings
Wears & Clothings
















