Technology tamfitronics
- Research Briefing
- Published:
Subjects
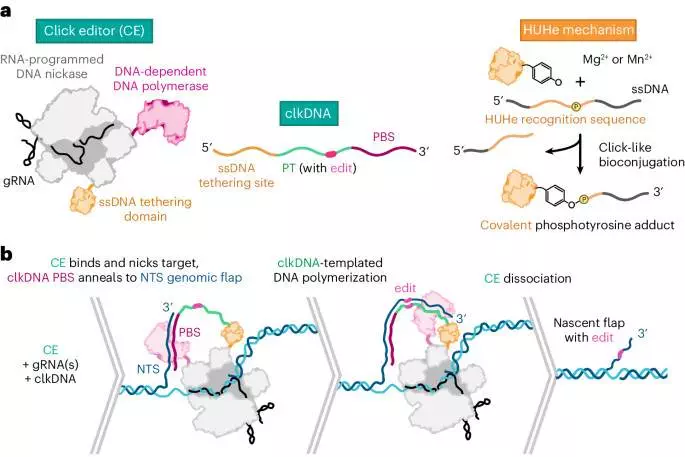
We developed click editors, comprising HUH endonucleases, DNA-dependent DNA polymerases and CRISPR–Cas9 nickases, which together enable programmable precision genome engineering from simple DNA templates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
214,86 € per year
only 17,91 € per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

References
-
Halperin, S. O. et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 550248–252 (2018). This manuscript describes the fusion of an error-prone polymerase to CRISPR–Cas9 to mutagenize specified genomic regions.
-
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576149–157 (2019). This paper reports the development of prime editing, a precise genome-editing approach that uses reverse transcriptases to write specific edits into a genome.
-
Levesque, S., Cosentino, A., Verma, A., Genovese, P. & Bauer, D. E. Enhancing prime editing in hematopoietic stem and progenitor cells by modulating nucleotide metabolism. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02266-4 (2024). This manuscript reports the dependence of RT-based genome editing technologies, such as prime editors, on cellular dNTP levels.
-
Aird, E. L. et al. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 154 (2018). This study describes the use of HUHes to recruit ssDNA templates to Cas9-mediated DNA breaks to improve homology-directed repair efficiencies.
-
Liu, B. et al. Targeted genome editing with a DNA-dependent DNA polymerase and exogenous DNA-containing templates. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01947-w (2023). This paper reports the development of DNA polymerase-based editors using RNA or DNA substrates for precise genome editing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Ferreira da Silva, J. et al. Click editing enables programmable genome writing using DNA polymerases and HUH endonucleases. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02324-x (2024).
Rights and permissions
About this article
Cite this article
Precise and versatile genome editing with click editors. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02340-x
-
Published:
-
DOI: https://doi.org/10.1038/s41587-024-02340-x